Epigenetics - An eye-opening role for Tet3
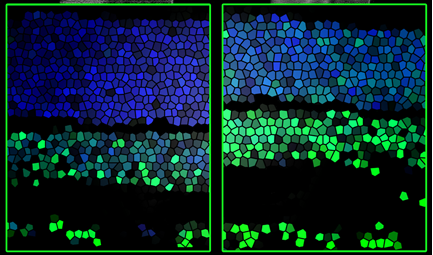 Retina sections: Note that the level of the modified DNA base 5hmC (green) increases from the second (left) to the third (right) post-natal week.
Retina sections: Note that the level of the modified DNA base 5hmC (green) increases from the second (left) to the third (right) post-natal week.
Vision is a highly complex process, and requires the construction of a correspondingly complicated functional network made up of diverse nerve-cell types in the developing retina. In mammals, these circuits are formed within a week after the eye’s first direct exposure to light. During this period, neuronal precursor cells differentiate into various types of mature nerve cells, which form specific patterns of functional connections with each other. The profound morphological and functional changes that occur during this maturation process require exquisitely precise control of the genetic programs that direct them. A joint project undertaken by LMU researchers led by PD Dr. Stylianos Michalakis and Professor Thomas Carell, both of whom are affiliated with the Center for integrated Protein Science Munich (CiPSM, a Center of Excellence), has now elucidated a previously unknown mechanism that is intimately involved in this phase of retinal development.
All the cells in multicellular organisms carry the same set of genes in their nuclear DNA. What distinguishes different cell types from each other is the subset of genes activated in each – and which genes are activated at any given time is in part regulated by patterns of chemical modifications (‘epigenetic marks’) of the nucleotide subunits in the genomic DNA. Alterations in these patterns play an essential role in controlling the maturation of the neuronal network during early post-natal development. One of the processes involved results in the attachment of methyl groups to specific nucleotides in the DNA. The methyl group serves as a label or tag, which is recognized by the machinery involved in controlling gene expression. In general, methylation leads to gene inactivation: The mark prevents the transcription of a particular section of the genomic DNA into molecules that direct protein synthesis. However, in 2009 it was discovered that the methylated form of the cytosine nucleotide can be further modified by enzymes of the Tet family. The resulting modified base, 5-hydroxymethylcytosine (5hmC), was already known but had been regarded as an intermediate in active demethylation. “Ever since 5hmC was shown to be the product of a targeted modification reaction, people have suspected that it might play a role in switching genes on and off,” says Michalakis. “At all events, mature nerve cells in the retina contain high levels of 5hmC in their nuclear DNA, and we hypothesized that the production of 5hmC by Tet enzymes plays a significant part in the development of the neuronal network. But it was unclear how the modification is carried out and what its effects are.”
The LMU team has now shown that the enzyme Tet3 interacts specifically with a DNA-bound transcriptional repressor called REST, and that this enables it to convert the repressive methylated epigenetic mark into the permissive 5hmC. Moreover, they found that Tet3 also participates in the next step in the transition of these genes from the inactive to the active state – by making them accessible to the transcriptional apparatus. It does this by binding to enzymes that attach methyl groups to the “packaging” proteins around which the DNA is wrapped. In this way, inactive DNA regions that were previously maintained in a condensed conformation and repressed by REST are opened up for copying to RNA.
The next question that arises is how, and under what circumstances, the Tet3-driven mechanism is itself activated. “We would also like to know whether defects in this mechanism might be responsible for certain diseases. If so, then our findings could also be of therapeutic use,” Michalakis adds.