Combined Use with Other Programs
The generation of Tinker input files for structures other
than peptides and nucleotides is most easily achievd through the combined use with other
programs. The following example uses GaussView and
MOLDEN to generate input files for geometry optimization
and conformational search with Tinker.
The example chosen here explores the conformational space of n-propylcyclohexane. This system
can easily be constructed from the fragments available in the GaussView
builder. After construction the molecule can be save as a standard Gaussian
input file such as "cyc1.com". Conversion of this file to the Tinker input
file format is most easily achieved by reading in the file with molden42 cyc1.com
and by writing out the structure as "cyc1.xyz" using the "Write" menu in Molden
specifying the Tinker file format. At this point the input file for the
system is as follows:
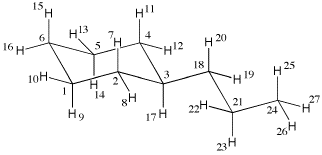
27 molden generated tinker .xyz (mm3 param.)
1 C 0.000000 0.000000 0.000000 1 2 6 9 10
2 C 0.000000 0.000000 1.515106 1 1 3 7 8
3 C 1.411078 0.000000 2.067037 1 2 4 17 18
4 C 2.215615 1.160661 1.517374 1 3 5 11 12
5 C 2.216276 1.160172 0.002249 1 4 6 13 14
6 C 0.805655 1.158876 -0.550551 1 1 5 15 16
7 H -0.545634 0.906562 1.887655 5 2
8 H -0.549859 -0.901910 1.890400 5 2
9 H 0.433029 -0.965385 -0.372688 5 1
10 H -1.054220 0.063502 -0.375597 5 1
11 H 1.780770 2.125272 1.889909 5 4
12 H 3.269752 1.098666 1.893397 5 4
13 H 2.765704 2.062286 -0.373071 5 5
14 H 2.762877 0.253849 -0.369606 5 5
15 H 0.299933 2.124191 -0.284547 5 6
16 H 0.840716 1.093938 -1.669099 5 6
17 H 1.916711 -0.964546 1.798419 5 3
18 C 1.364377 0.086165 3.603915 1 3 19 20 21
19 H 2.361016 0.065608 3.992744 5 18
20 H 0.886440 0.997954 3.895664 5 18
21 C 0.571131 -1.110379 4.161271 1 18 22 23 24
22 H -0.476441 -0.898305 4.111106 5 21
23 H 0.786626 -1.982818 3.580485 5 21
24 C 0.975456 -1.356491 5.626723 1 21 25 26 27
25 H 0.839215 -0.457118 6.190166 5 24
26 H 0.364882 -2.132066 6.039741 5 24
27 H 2.003197 -1.651289 5.668466 5 24
|
| 
|
The first line contains the number of atoms in the input file (here 27) together with a
comment (here stating that the structure has been generated with molden). The second
line specifies the first center with running number "1", being of atom type "C" (a carbon atom),
followed by the xyz coordinates, the first atom being located at the origin of the
coordinate system in this particular case. The sixth column specifies the MM3 force field
atom type (here 1) and (through a maximum of four additional n umbers) the connectivity of the
current center. Center 1 is in this case connected to centers 2 and 6 (two carbon atoms)
and to centers 9 and 10 (two hydrogen atoms). It is clear from this connectivity that
atom 1 must be one of the ring carbon atoms in the cyclohexane ring. The other centers
are defined in an analogous manner.
Before being able to explore the conformational space of the system we still need to generate
an appropriate keyword file "cyc1.key" defining the force field to be used for the present study.
The most appropriate force field for hydrocarbons and related organic molecules ist the MM3 force
field and a minimal keyword file will accordingly be:
parameters /usr/local/tinker/params/mm3
This line points to the MM3 parameter file that can also be inspected here.
The atom types 1 and 5 are described at the very beginning of the parameter file as
belonging to sp3 hybridized carbon atoms as present in alkanes and as hydrogen
atoms connected to carbon atoms:
atom 1 C "CSP3 ALKANE" 6 12.000 4
atom 2 C "CSP2 ALKENE" 6 12.000 3
atom 3 C "CSP2 CARBONYL" 6 12.000 3
atom 4 C "CSP ALKYNE" 6 12.000 2
atom 5 H "EXCEPT ON N,O,S" 1 1.008 1
Searching the conformational space can again be achieved with the scan
program from the Tinker program suite. An appropriate command file
named "cyc1.run" including some sorting and cleanup is as follows:
scan cyc1 0 5 100 0.0001
grep "Map" cyc1.log | cat > tempo
sort tempo -nr -k 6,6n -o cyc1.list
rm tempo
After making the file executable this command file can be executed with:
./cyc1.run >& cyc1.log &
Conformational search with the MM3 force field gives in this particular case rise to 7 different
conformers with the following MM3 force field energies (in kcal/mol):
Potential Surface Map Minimum 1 12.2568
Potential Surface Map Minimum 3 12.9967
Potential Surface Map Minimum 6 13.2390
Potential Surface Map Minimum 4 14.8350
Potential Surface Map Minimum 7 15.1229
Potential Surface Map Minimum 2 15.8632
Potential Surface Map Minimum 5 15.9882
The structures of these conformational minima are located in a series of files called
"cyc1.001" . . . "cyc1.007". A combined coordinate file can be generated from
these single structure files using the
archive
program of the Tinker program suite, which works quite
selfexplanatory. Using the UNIX command "cat" it is also possible to combine the
single structure files into one archive, now also reflecting the sequence of
relative stability:
cat cyc1.001 cyc1.003 cyc1.006 cyc1.004 cyc1.007 cyc1.002 cyc1.005 > cyc1.arc
The single structure files as well as the archive files generated with archive
or the cat command can be read with Molden and
converted from there again to input files for quantum mechanical calculations with
Gaussian. On analyzing the results obtained for n-propylcyclohexane you will
realize that the conformers differ in the orientation of the n-propyl side chain only. This is due to
the conformational search algorithm used in scan.
last changes: 18.11.2005, HZ
questions & comments to: zipse@cup.uni-muenchen.de
