Construction and Optimzation of Peptides
Building up a peptide is achieved using the protein subprogram in either an
interactive mode (when the program will prompt you for input) or by putting the parameters into an input
file and calling the program together with the input file through a shell script. A list of amino acids
and their abbreviations can be found here. The input file (termed here
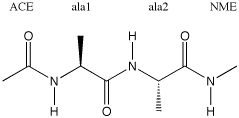
testz1.dat) for building up the dialanine tripeptide with N-terminal acetyl and C-terminal N-Me
amide capping groups is as follows:
testz1
ACE-(ala)2-NME
ACE
ala
ala
NME
n
|
| 
|
The first of these lines specifies the output file (here: testz1), the second line a title in
free format (here: ACE-(ala)2-NME), the third line the N-terminal residue, here taken to be a capping
acetyl group (ACE), the fourth and fifth lines specify the two alanine residues without any additional
parameters, and the sixth line the C-terminal N-methyl amide capping group (NME). One empty line terminates
the definition of the amino acid sequence. After specifying a non-cyclic peptide (n) the definition of the
peptide is finished. The input file described above can be executed simply from the command
line:
protein < testz1.dat
The above example assumes a matching parameter file (testz1.key) to exist containing additional information
such as the force field type and parameters to use. In case a matching keyword file does not (yet) exist,
the protein subprogram prompts the user for the corresponding information.
In case we want to use the amber99 parameters for the AMBER force field, the testz1.key file
is quite simple:
parameters /usr/local/tinker/params/amber99
This file is also needed in order to read the newly generated testz1.xyz file into
FFE.
The example chosen above generates a peptide structure in which all phi angles are set to -135.0 degrees,
all psi angles to +135.0 degrees, and all omega angles to 180.0 degrees. This generates an approximately
linear peptide chain found as substructures in beta sheets. The definition of these angles is shown below
for the ala-ala dipeptide, carrying an acetyl protecting group at the N-terminal end, and capping the
C-terminal end as N-methyl amide:
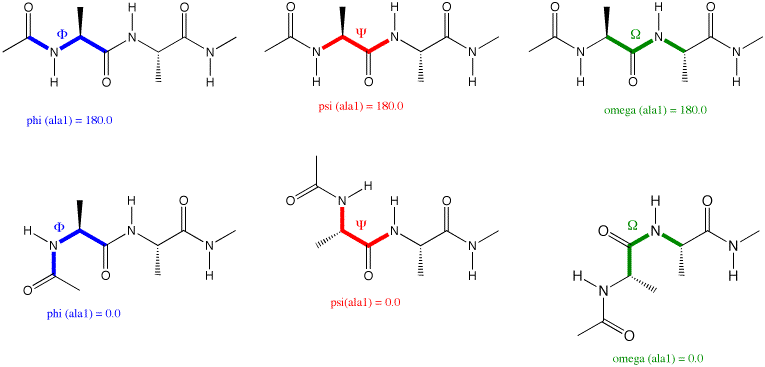
The value of omega (the dihedral angle describing rotation around the peptide bond) is often very
close to 180.0 degrees (a trans-peptide bond). In some selected cases, however, a cis-peptide bond
(with values for omega around 0.0 degrees) can also be found in protein structures. To which of the
two amino acids participating in a peptide bond the omega angle is attributed is a matter of personal
choice. In the Tinker input file the omega angle given for amino acid i
defines the peptide bond between amino acid
i and i+1. The values of phi and psi depend on the particular folding motive of the protein and the
following typical cases are often observed: (a) a right-handed helical turn (alpha helix) with
phi = -55.0 and psi = -45.0 degrees; (b) beta sheet structures are formed by extended chains with
phi = -135.0 and psi = +135.0 degrees (approximate values).
The angle information can be included in the protein
input file on the same line as the residue itself.
The following example shows the construction of the end-capped alanine decapeptide in a conformation with
phi and psi angles equal to -60.0 degrees, generating a rough starting conformation for a partial
alpha helix. Due to the presence of the two capping groups, omega angles can be defined for both the
leading and the tailing alanine residue:
testz1
testz1 ACE-(ala)10-NME, alpha helix input
ACE
ala -60.0 -60.0 180.0
ala -60.0 -60.0 180.0
ala -60.0 -60.0 180.0
ala -60.0 -60.0 180.0
ala -60.0 -60.0 180.0
ala -60.0 -60.0 180.0
ala -60.0 -60.0 180.0
ala -60.0 -60.0 180.0
ala -60.0 -60.0 180.0
ala -60.0 -60.0 180.0
NME
n
The xyz-coordinate files generated with protein can be minimzed with a number of
different optimization routines. Optimization of the peptide structure to a loose convergence criterion
of around 0.01 kcal/mol/angstrom can be achieved with the minimize program,
which is also known to work well with larger protein structures. As input the program requires the principal
name of the system (here: testz1) and the convergence criterion:
minimize testz1 0.01
The output consists of a new xyz-coordinate file as well as some information concerning the convergence
of the optimization process. In order to direct the latter into a log file, the construction of the
peptide and its optimization with minimize can be combined in one shell script (e.g. testz1.run):
protein < testz1.dat
minimize testz1 0.01
This short script can be executed (after making it executable with "chmod u+x testz1.run") with
the following command:
./testz1.run > testz1.log
The alanine decapeptide system can also be used to illustrate the performance of the various optimizers
such as minimize, newton, and
optimize contained in Tinker. The table below
shows the overall CPU times, cycle numbers, and final energy of the optimized structure for various
optimizers for convergence criteria of 0.01 and 0.001 kcal/mol/angstrom, respectively:
| optimizer | timea,c
(sec) | cyclesa | final energya
(kcal/mol)
| | timeb,c
(sec) | cyclesb | final energyb
(kcal/mol)
|
|---|
| minimize | 13.79 | 1099 | -18.9569 | | 15.52 | 1255 | -18.9574
|
| newton | 4.62 | 31 | -18.6278 | | 5.18 | 32 | -18.6278
|
| optimize | 34.38 | 1499 | -10.6792 | | 35.03 | 1521 | -10.6792
|
aconvergence criterion of 0.01 kcal/mol/angstroms; bconvergence criterion of
0.001 kcal/mol/angstroms; call times have been obtained using the UNIX time
command
We can see, that for small systems such as the one chosen here, the newton
optimizer is much more efficient, the difference to the minimize optimizer
being increasingly large with increasing convergence criteria. It can also be seen that the newton
optimizer performs a small number of expensive steps, while the minimize
optimizer uses a very larger
number of cheap steps. While these two optimizers converge onto essentially the same final structure,
this is not the case with the optimize optimizer. The latter is not only
the slowest optimizer, but also converges onto a much less favorable structure. These findings are,
however, only relevant for small systems such as the alanine decapeptide chosen here.
last changes: 03.11.2004, HZ
questions & comments to: zipse@cup.uni-muenchen.de


